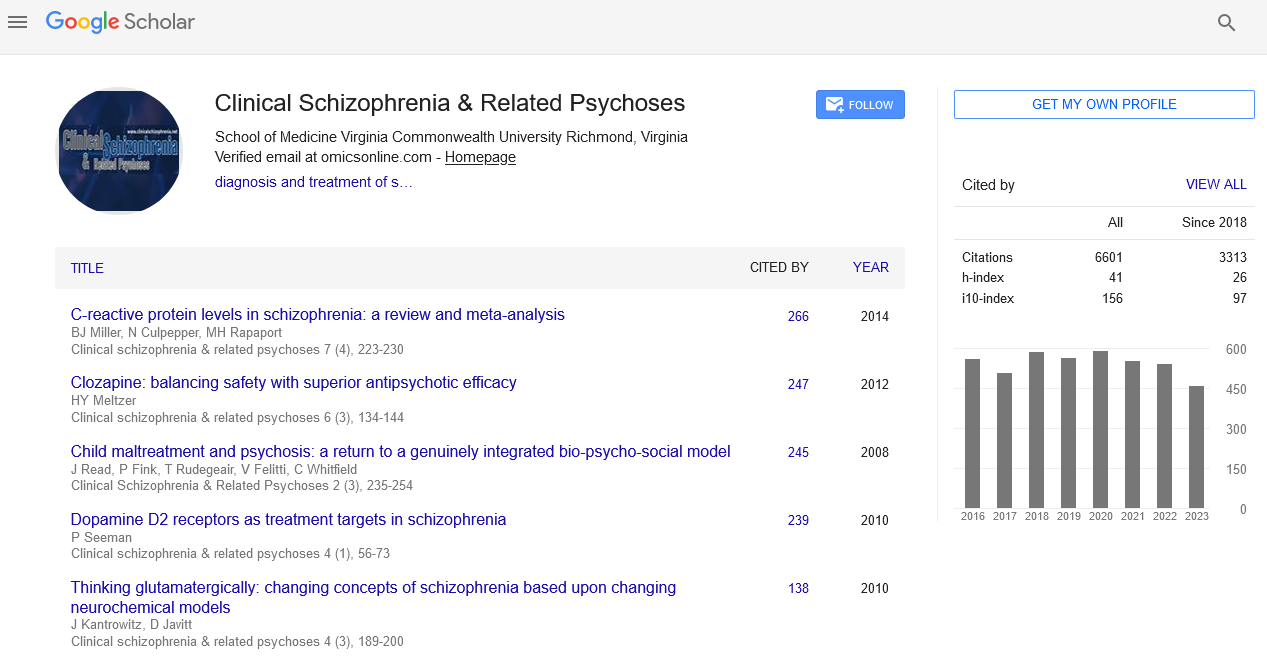
Abstract
Colchicine, Baricitinib, Efornithine, Umifenofir, Hydroxychloroquine, Azathioprine, Cycloserine and Linoleic Acid to Stratify Synergistic Responses on SARSCOV-2 Main 6lu7 Protease: Quantum Mechanics Driven Applications of Artificial Deep Learning Similarities for a NOS3 Hypoxic Based Drug Retargeting Methodology to Treat COVID-19
Author(s): John-Ioannis GrigoriadisSARS coronavirus 2 (SARS-CoV-2) of the family Coronaviridae is an enveloped, positive-sense, single-stranded RNA betacoronavirus encoding a SARS-COV-2 Main protease PDB:6LU7 with Unliganded Active Site (2019-NCOV, Coronavirus Disease 2019, that infect humans historically. Hydroxychloroquine (HCQ), an antimalarial has been proposed as possible treatment for coronavirus disease-2019 (COVID-19). Ischemic heart disease (IHD) is the leading cause of death and a major economic burden worldwide. It is the cause of over 30% of total annual deaths and constitutes 17% of overall national health expenditure in the United States (U.S.) The single nucleotide polymorphism (SNP) NOS3 894GT located in exon 7 (also known as Glu298Asp, rs1799983) is a genetic marker that has been specifically linked to an increased risk of IHD, hypertension, coronary spasms, and stent re-stenosis. Quantum mechanics, molecular mechanics, molecular dynamics (MD), and combinations have shown superior performance to other drug design approaches providing an unprecedented opportunity in the rational drug development fields and for the developing of innovative drug repositioning methods. The availability of newer modeling techniques with integration of the state of art deep learning algorithm can be modeled as a recommendation system that recommends novel treatments based on known drug-disease powerful computational resources. The formulation under this drug repositioning recommendation system could provide us with a deep learning model and generate the target-focused de novo libraries for the generations of a generate good-quality data and reliable predictions for new chemical entities, impurities, monoclonal antibodies, chemicals, natural products, and a lot of other substances fuelling further development and growth of the field to balance the trade-off between the molecular complexity and the quality of such predictions assuming that the hidden factors that cannot be obtained by any other method where new drug-disease associations having not been validated can now be screened. Drug repurposing offers a promising alternative by integrating related data sources to dramatically shorten the process of traditional de novo development of a drug. We here present an approach of a fast Singular Constructed Classification and Regression NOS3 894GT –SARS-COV2-ORF-1a Model which could be subsequently used for virtual screening against the generated de novo cluster of COVID19 libraries and diverse FDA chemical libraries. QMMM Quantum Deep Learning functional Value Thresholding (SVT) algorithm to prioritize drug combinations in high-throughput screens and to stratify synergistic responses on SARS-COV-2 Main protease PDB:6LU7 With Unliganded Active Site (2019-NCOV, Coronavirus Disease 2019, by co-targeting the NOS3 894GT mutation for medications to treat COVID-19. At the core of our approach is the observation that the likelihood of synergy increases when screening small molecule, anti-viral compounds and other FDAs with either strong functional pharmacophoric similarity or dissimilarity. In this research paper, we estimated the druggable similarity by applying an inverse docking multitask machine learning approach to basal gene expression in acute respiratory distress syndrome and response to single drugs. We tested 18 small molecules and predicted their synergies in COVID19 SARS-COV-2 main protease PDB:6LU7 with unliganded active site (2019-NCOV), which is associated with 1,000,000 deaths worldwide, to devise therapeutic strategies, repurpose existing ones in order to counteract highly pathogenic SARS-CoV-2 infection and the associated NOS3-COVID-19 pathology. We anticipate that our approaches can be used for prioritization of drug combinations in large scale screenings, and to maximize the efficacy of the Colchicine, Baricitinib, Efornithine, Umifenofir, Hydroxychloroquine, Azathioprine, Cycloserine, Cobicistat and Linoleic acid drugs already known to induce synergy, ultimately enabling COVID19 hypoxic patient stratification.